Search Your Topic
DNA Replication-Lecture-1
DNA Replication: the process of DNA Synthesis
Purpose of replication
- Basis for inheritance
- The fundamental process occurring in all cells for copying DNA to transfer the genetic information to daughter cells
- Each cell must replicate its DNA before division.
Salient features of DNA replication
1) Semi conservative
- Parental strands are not degraded
- Base pairing allows each strand to serve as a template for a new strand
- New duplex is 1/2 parent template & 1/2 new DNA (figure-1)

Figure-1- Semiconservative mode of replication
2) Semi discontinuous- Leading & Lagging strands
- Leading strand – continuous synthesis
- Lagging strand – Okazaki fragments joined by ligases (figure-2)

Figure-2- Semi- discontinuous type of replication
3) The energy of Replication- The nucleotides arrive as nucleoside triphosphates with their own energy source for bonding
- DNA base, sugar with PPP
- P-P-P = energy for bonding
- Mono phosphate form is bonded by enzyme DNA polymerase III, a pyrophosphate is released, which is further broken down, the energy released is used for polymerization.
GTP———————-> GMP +PPi
4) Template reading and direction of polymerization
- Each DNA strand serves as a template to guide the synthesis of the complementary strand.
- The template is read in the 3′-5′ direction while polymerization takes place in the 5′-3′ direction.
5) Primer is needed
The DNA polymerase can only add nucleotides to 3′ end of a growing DNA strand needs a “starter” nucleotide to make a bond
- Primer serves as a starter sequence for DNA polymerase III
- RNA primer is synthesized by Primase
- RNA Primer has a free 3’OH group to which the first Nucleotide is bound (figure-3)
- Only one RNA Primer-required for the leading strand
- RNA Primers for the lagging strand depend on the number of “OKAZAKI FRAGMENTS”
Figure 3- The deoxyribonucleotides are attached to the 3’OH group of the primer by 3’-5’ phosphodiester linkage.
DNA Replication (Prokaryotes) Steps
- Identification of the origins of replication
- Unwinding (denaturation) of dsDNA to provide an ssDNA template
- Formation of the replication fork
- Initiation of DNA synthesis and elongation
- Primer removal and ligation of the newly synthesized DNA segments
- Termination of replication
Components of Replication
Enzymes
- DNA polymerases- Deoxynucleotide polymerization (I, II, and III)
- Helicase -Processive unwinding of DNA
- Topoisomerases (I and II) relieve the torsional strain that results from helicase-induced unwinding
- RNA primase initiates the synthesis of RNA primers
- DNA ligase-seals the single strand nick between the nascent chain and Okazaki fragments on the lagging strand
Proteins
- dna A protein for identification of the origin of replication
- Single-strand binding proteins prevent premature reannealing of dsDNA and to protect from attack by nucleases
- Deoxyribonucleotides for polymerization
Step-1- Identification of Origin of Replication
- At the origin of replication (ori), there is an association of sequence-specific dsDNA-binding proteins with a series of DNA sequences.
- In E Coli, the oriC is bound by the protein dnaA (figure-4)
- A complex is formed consisting of 150–250 bp of DNA and multimers of the DNA-binding protein.
- This leads to the local denaturation and unwinding of an adjacent A+T-rich region of DNA.
Step-2- Unwinding of double-stranded DNA to provide a single-stranded template
- The interaction of proteins with ori defines the start site of replication and provides a short region of ssDNA essential for initiation of synthesis of the nascent DNA strand.
- DNA Helicase allows for the processive unwinding of DNA.
- Single-stranded DNA-binding proteins (SSBs) stabilize this complex.
- Replication occurs in both directions along the length of DNA and both strands are replicated simultaneously.
- This replication process generates “replication bubbles” (figure-4)
- A further unwinding of DNA creates a replication fork.

Figure-4- Identification of the origin of replication and formation of a replication bubble.
Step-3 Formation of Replication Fork-
A replication fork consists of four components that form in the following sequence:
- DNA helicase unwinds a short segment of the parental duplex DNA
- A primase initiates the synthesis of an RNA molecule that is essential for priming DNA synthesis;
- DNA polymerase initiates nascent, daughter strand synthesis; and
- SSBs bind to ssDNA and prevent premature reannealing of ssDNA to dsDNA (figure-5)
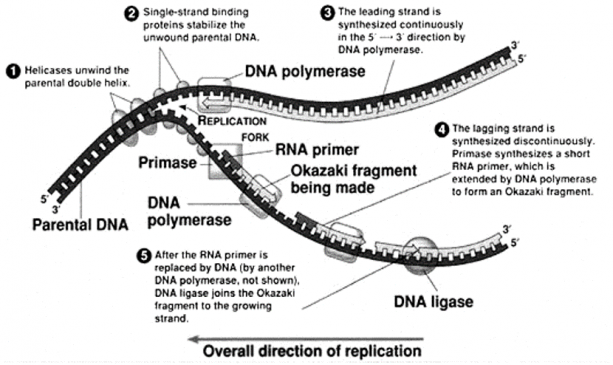
Figure-5 – components and the processes involved at the replication fork.
