Search Your Topic
DNA Damage and repair
DNA in the living cell is subjected to many chemical alterations. If the genetic information encoded in the DNA is to remain uncorrupted, any chemical changes must be corrected. A failure to repair DNA produces a mutation.
Agents that Damage DNA
- Radiations- Highly reactive oxygen radicals produced during normal cellular respiration as well as by other biochemical pathways
- Ionizing radiation such as gamma rays and x-rays
- Ultraviolet rays, especially the UV-C rays (~260 nm) that are absorbed strongly by DNA but also the longer-wavelength UV-B that penetrates the ozone shield
- Chemicals in the environment
- many hydrocarbons, including some found in cigarette smoke
- some plant and microbial products, e.g. the Aflatoxin produced in moldy peanuts
- Chemicals used in chemotherapy, especially chemotherapy of cancers.
Types of DNA damage
DNA in cells suffers a wide range of damage:
- Purine bases are lost by the spontaneous fission of the base-sugar link;
- Cytosines, and occasionally adenines, spontaneously deaminate to produce Uracil and hypoxanthine respectively
- Many chemicals, for example, Alkylating agents, form adducts with DNA bases
- Reactive oxygen species in the cell attack purine and pyrimidine rings
- Errors in DNA replication result in incorporation of a mismatched base
- Ionizing radiation causes single- or double-strand breaks;
- Errors in replication or recombination leave strand breaks in DNA.
- Crosslinks- Covalent linkages can be formed between bases on the same DNA strand (“intrastrand”) or on the opposite strand (“interstrand”). Ultraviolet light causes adjacent thymines to form a stable chemical dimer
All these lesions must be repaired if the cell has to survive. The importance of effective DNA repair systems is highlighted by the severe diseases affecting people with deficient repair systems
DNA Repair
To cope with all these forms of damage, cells must be capable of several different types of DNA repairs. DNA repair seldom involves simply undoing the change that caused the damage. Almost always a stretch of DNA containing the damaged nucleotide(s) is excised and the gap is filled by resynthesis.
DNA repair can be grouped into two major functional categories:
A) Direct Damage reversal
B) Excision of DNA damage
A) Direct Damage Reversal
The direct reversal of DNA damage is by far the simplest repair mechanism that involves a single polypeptide chain, with enzymatic properties that binds to the damage and restores the DNA genome to its normal state in a single reaction step. The major polypeptides involved in this pathway are:
i) DNA photolyases, the enzymes responsible for removing cyclobutane pyrimidine dimers from DNA in a light-dependent process called photoreactivation (figure-1).

Figure-1- showing the mechanism of direct reversal of DNA damage by photolyases
ii) O6-methylguanine-DNA methyltransferase I and II (MGMT), also called DNA-alkyltransferase, remove the modified bases like O6-alkylguanine and O4-alkylthymine.
The photolyase protein is not found in all living cells. However, DNA-alkyltransferases are widespread in nature. Some of the drugs used in cancer chemotherapy also damage DNA by alkylating. Some of the methyl groups can be removed MGMT enzyme; however, the enzyme can only do it once, so the removal of each methyl group requires another molecule of enzyme.
B ) Excision of DNA damage – includes
a) Base excision repair (BER)
b) Nucleotide excision repair (NER),
c) Mismatch repair (MMR) and
d) Strand break repairs.
In these reactions, a nucleotide segment containing base damage, double-helix distortion or mispaired bases is replaced by the normal nucleotide sequence in a new DNA polymerase synthesis process. All of these pathways have been characterized in both bacterial and eukaryotic organisms.
i) Base excision repair (BER)- figure-2
BER is initiated by DNA glycosylases, which catalyze the hydrolysis of the N-glycosidic bonds, linking particular types of chemically altered bases to the deoxyribose-phosphate backbone. Thus, DNA damage is excised as free bases, generating sites of base loss called apurinic or apyrimidinic (AP) sites. Another means of AP site generation is the depurination or depyrimidation of DNA, due to the spontaneous hydrolysis of N-glycosidic bonds. The AP sites are substrates for AP endonucleases (Figure-2). These enzymes produce incisions in duplex DNA as a result of the hydrolysis of a phosphodiester bond immediately 5′ or 3′ to each AP site. The ribose-phosphate backbone is then removed from the DNA through the action of a specific exonuclease called deoxyribophosphodiesterase or dRpase. Finally, the DNA polymerase and a ligase catalyze the incorporation of a specific deoxyribonucleotide into the repaired site, enabling correct base pairing (Figure-2).

Figure-2- showing the mechanism of the base excision repair system.
ii) Nucleotide excision repair (NER)
Several types of agents generate bulky base adducts in DNA, leading to a significant distortion of the DNA helix. The most widely studied of these DNA damaging agents is UV radiation, responsible for thymine dimers, which produce a bend of ~30° in the DNA. Some chemical agents form DNA cross-links, which are particularly hazardous. These cross-links produce conformational distortions in DNA; they are substrates for DNA endonucleases that make an incision in DNA, several nucleotides to each side of the damage, generating a potential oligonucleotide fragment. Subsequent helicase reactions promote the excision of this fragment. The resulting gap is filled by DNA polymerase synthesis and covalently sealed by DNA ligase. These sequential enzymatic reactions, initiated by a specific endonuclease that recognizes the DNA distortion, are called NER (Nucleotide excision repair) (figure-3).

Figure-3- showing the mechanism of nucleotide excision repair
NER is a much more complex biochemical process than BER, especially in eukaryotic cells. Several gene products are required in a multiple-step process, during which the ordered assembly of DNA proteins provides an enzymatic complex that discriminates damaged from undamaged DNA.
In Escherichia coli, there are three specific proteins, called UvrA, B and C, involved in lesion recognition and endonuclease incision. This fragment is released by UvrD helicase action, generating a gap that is finally submitted to repair synthesis (figure-4).

Figure-4- showing mechanism of nucleotide excision repair in E.Coli
Transcription-Coupled NER
Nucleotide-excision repair proceeds most rapidly
- in cells whose genes are being actively transcribed
- on the DNA strand that is serving as the template for transcription.
If RNA polymerase II, tracking along with the template (antisense) strand), encounters a damaged base, it can recruit other proteins, e.g., the CSA and CSB proteins, to make a quick fix before it moves on to complete transcription of the gene.
iii) Mismatch repair (MMR)
Mismatch repair corrects errors made when DNA is copied. For example, a C could be inserted opposite an A, or the polymerase could slip or stutter and insert two to five extra unpaired bases. Specific proteins scan the newly synthesized DNA, using adenine methylation within a GATC sequence as the point of reference (Figure -5). The template strand is methylated, and the newly synthesized strand is not. This difference allows the repair enzymes to identify the strand that contains the errant nucleotide which requires replacement. If a mismatch or small loop is found, a GATC endonuclease cuts the strand bearing the mutation at a site corresponding to the GATC. An exonuclease then digests this strand from the GATC through the mutation, thus removing the faulty DNA. This can occur from either end of the defect is bracketed by two GATC sites. This defect is then filled in by normal cellular enzymes according to base-pairing rules (figure-5).

Figure-5- showing the mechanism of mismatch repair system
In E coli, three proteins (Mut S, Mut L, and Mut H) are required for recognition of the mutation and nicking of the strand. Other cellular enzymes, including ligase, polymerase, and SSBs, remove and replace the strand. The process is somewhat more complicated in mammalian cells, as about six proteins are involved in the first steps (figure-6).
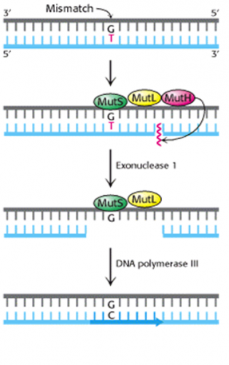
Figure-6-showing mechanism of mismatch repair system in E. coli
Faulty mismatch repair has been linked to hereditary nonpolyposis colon cancer (HNPCC), one of the most common inherited cancers.
B) Repairing Strand Breaks
Ionizing radiation and certain chemicals can produce both single-strand breaks (SSBs) and double-strand breaks (DSBs) in the DNA backbone.
i) Single-Strand Breaks (SSBs)
Breaks in a single strand of the DNA molecule are repaired using the same enzyme systems that are used in Base-Excision Repair (BER).
ii) Double-Strand Break Repair
There are two mechanisms by which the cell attempts to repair a complete break in a DNA molecule:
1) The direct joining of the broken ends. This requires proteins that recognize and bind to the exposed ends and bring them together for ligating. This type of joining is also called Nonhomologous End-Joining (NHEJ). A protein called Ku is essential for NHEJ.(figure-7)
Errors indirect joining may be a cause of the various translocations that are associated with cancers. Examples:
- Burkitt’s lymphoma
- Philadelphia chromosome in chronic myelogenous leukemia (CML)
- B-cell leukemia
2) Homologous Recombination. Here the broken ends are repaired using the information on the intact (figure-7).
- sister chromatid, or on the
- homologous chromosome
- same chromosome if there are duplicate copies of the gene on the chromosome oriented in opposite directions (head-to-head or back-to-back).
Two of the proteins used in homologous recombination are encoded by the genes BRCA1 and BRCA2. Inherited mutations in these genes predispose women to breast and ovarian cancers.

Figure-7-showing mechanism of double-strand break repair
Meiosis also involves DSBs
Recombination between homologous chromosomes in meiosis I also involve the formation of DSBs and their repair. Meiosis I with the alignment of homologous sequences provides a mechanism for repairing damaged DNA.
Diseases associated with defective DNA repair system
Some of the examples include:
- Ataxia telangiectasia
- Bloom syndrome
- Cockayne’s syndrome
- Progeria (Hutchinson-Gilford Progeria syndrome)
- Rothmund-Thomson syndrome
- Trichothiodystrophy
- Werner syndrome
- Xeroderma pigmentosum
- Hereditary non-polyposis colon cancer.
